Do not directly compare cross-validation results between genomic and phenomic predictions
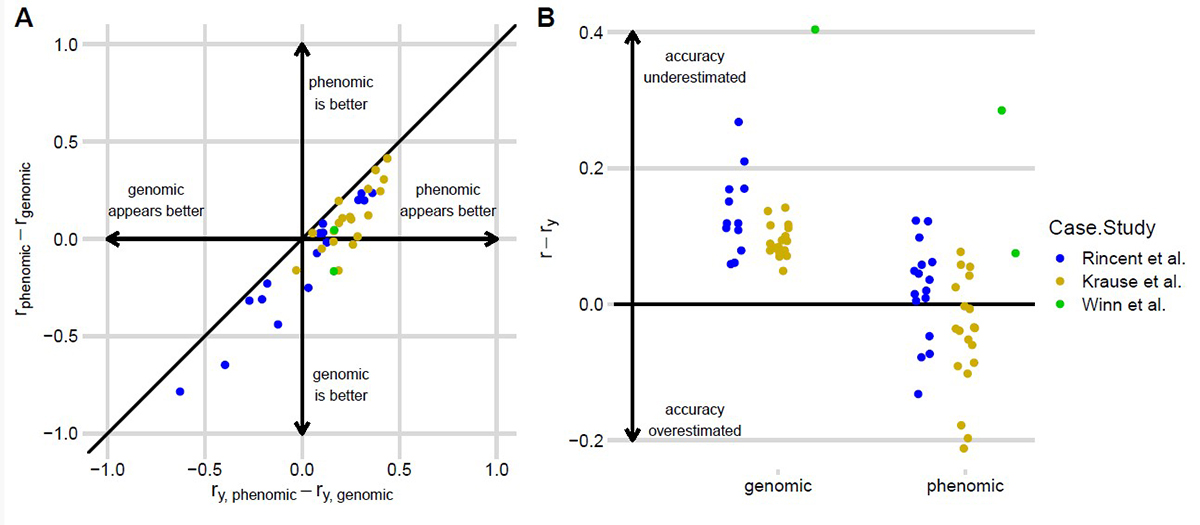
About the figure above: Predictive ability overestimates the relative accuracy of phenomic prediction (PP) versus genomic prediction (GP) models. Colored dots correlate with different traits tested in each case study. (A) Differences between estimated PP and GP accuracies are plotted against their estimated predictive abilities. Points to the right of the line are where the accuracy of these PP models tends to be overestimated compared with GP models, even when the PP models are actually less accurate (lower right quadrant). (B) Differences between estimated accuracies and predictive abilities for GP and PP models, binned by case study. Figure courtesy of Fangyi Wang, Mitchell J. Feldmann, and Daniel E. Runcie.
Crop breeders use both genotypic and phenotypic data to select the best candidates for new crop lines. Over the past two decades, genomic selection, a method to estimate a plant's potential performance using genetic information, has proven to be a powerful method for making selection decisions. Recently, breeders have begun to collect high-throughput phenotyping data as well, a technique where technology aids in collecting a large amount of data on complex plant traits, non-destructively. This method can also be used to improve selection decisions and is often thought of as more efficient and less expensive than genotype data. Both types of data require complex statistical models, and both are typically evaluated using cross-validation values to estimate the accuracy of each model.
While breeders need both types of models to have high accuracy, researchers at the University of California, Davis identified a limitation of using cross-validation to compare the accuracy of these models. Specifically, accuracy means something completely different when describing a phenotypic prediction model vs. a genomic prediction model. When used to evaluate genomic prediction models, cross-validation estimates the accuracy of predicted breeding values, which are values that describe the genetic potential of individuals to pass down desirable traits to the next generation. In contrast, when used to evaluate phenotypic prediction models, cross-validation estimates the accuracy of predicted phenotypic values of the candidates themselves. Both predictions are useful for breeders, but at different stages of a breeding program. Therefore, comparing them is like “comparing apples to oranges” and doesn’t help evaluate whether either type of model is useful.
The researchers showed that by directly comparing estimated genotypic prediction accuracy to phenotypic prediction accuracy, previous studies have mistakenly concluded that phenotypic prediction was more useful. Instead, the researchers concluded that these technologies are complementary and should be evaluated separately.
Dig deeper
Wang, F., Feldmann, M. J., & Runcie, D. E. (2025). Do not benchmark phenomic prediction against genomic prediction accuracy. The Plant Phenome Journal, 8, e70029. https://doi.org/10.1002/ppj2.70029
Text © . The authors. CC BY-NC-ND 4.0. Except where otherwise noted, images are subject to copyright. Any reuse without express permission from the copyright owner is prohibited.












