Nondestructive Spectroscopy of Kernels Helps Predict Maize Agronomic Traits
To evaluate and identify stable, high‐performing crops, many commercial and research breeding programs implement genomic prediction where DNA sequence data are extracted from different varieties and used in downstream statistical analysis. However, nondestructive methods of obtaining data for prediction of crop performance could save time and costs.
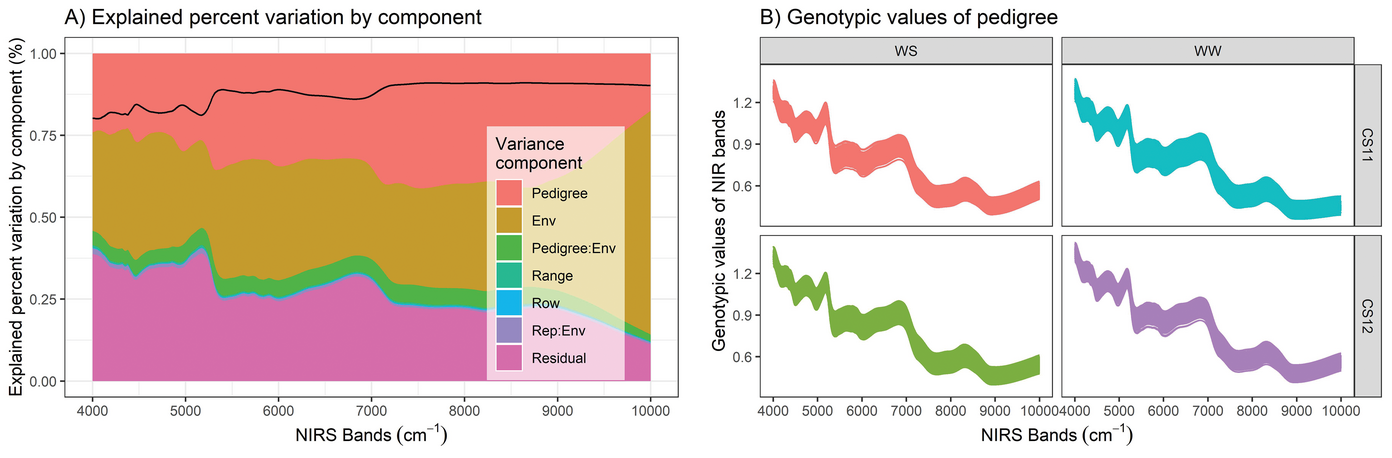
To explore this, researchers at Texas A&M University used near‐infrared reflectance spectroscopy (NIRS) to scan maize kernels from four distinct growing environments and recorded reflected light (over 3,000 wavelengths). By using a similar statistical analysis to what is used for handling large genomic data sets, the researchers were able to test how well NIRS‐based prediction performed vs. genomic prediction. Though in several instances genomic prediction outperformed NIRS‐based prediction, they found that NIRS performed comparably in across‐environment prediction.
These findings are important for breeding programs seeking to screen varieties at scale and nondestructively by harnessing information from intact maize kernels. High‐throughput methods such as NIRS have the potential to accelerate the pace of progress for variety improvement and can complement or act as a standalone method for prediction of performance.
Adapted from
DeSalvio, A. J., Adak, A., Murray, S. C., Jarquín, D., Winans, N. D., Crozier, D., & Rooney, W. L. (2024). Near-infrared reflectance spectroscopy phenomic prediction can perform similarly to genomic prediction of maize agronomic traits across environments. The Plant Genome, e20454. https://doi.org/10.1002/tpg2.20454
Text © . The authors. CC BY-NC-ND 4.0. Except where otherwise noted, images are subject to copyright. Any reuse without express permission from the copyright owner is prohibited.