Graph pangenomes support bread wheat breeding
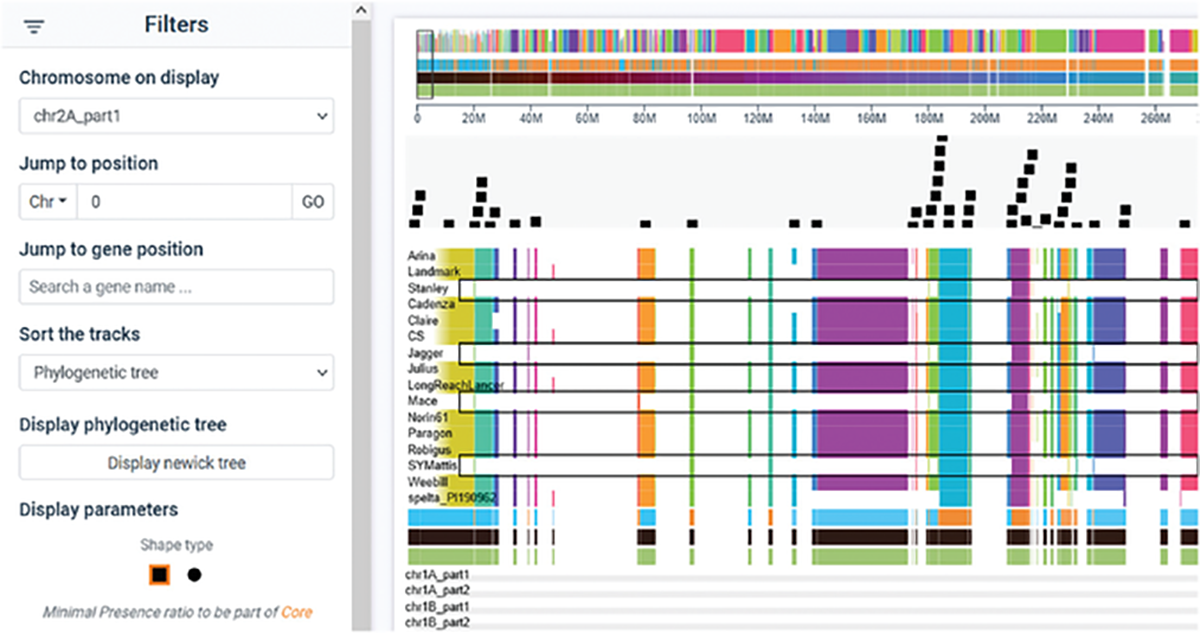
Bread wheat is one of humanity’s staple foods, but climate change threatens yields. Genomics methods can accelerate breeding of advanced wheat varieties, but the wheat genome is very large and complex and differs significantly among varieties. Graph pangenomes are a new type of genome reference that represents the diversity of a species rather than the gene content of an individual. Using a graph pangenome as a reference can help capture missing heritability when associating traits with DNA sequence variation, information valuable for crop breeding.
In a recent article in The Plant Genome, a research team presents a wheat graph pangenome combining the assemblies of 16 bread wheat cultivars. They show that only 65% of the genomic areas are present in all cultivars with several agronomically important genes being highly variable. This graph is visualized using Panache, an online tool for visualizing and exploring linear representations of pangenomes. The wheat Panache database allows users to search the assembly for regions of interest to assess the presence or absence of genes. The graph pangenome and wheat Panache will help breeders and researchers identify variation in the genome responsible for important traits, which can be used for breeding advanced, highly productive wheat cultivars.
Dig deeper
Bayer, P.E., Petereit, J., Durant, É., Monat, C., Rouard, M., Hu, H., … & Edwards, D. (2022). Wheat Panache: A pangenome graph database representing presence–absence variation across sixteen bread wheat genomes. The Plant Genome, e20221. https://doi.org/10.1002/tpg2.20221
Text © . The authors. CC BY-NC-ND 4.0. Except where otherwise noted, images are subject to copyright. Any reuse without express permission from the copyright owner is prohibited.











